University of Delhi
Advancing Drug Development: Integrating Computational Approaches to Combat Infections and Cancer at Dr. B. R. Ambedkar Center for Biomedical Research, University of Delhi
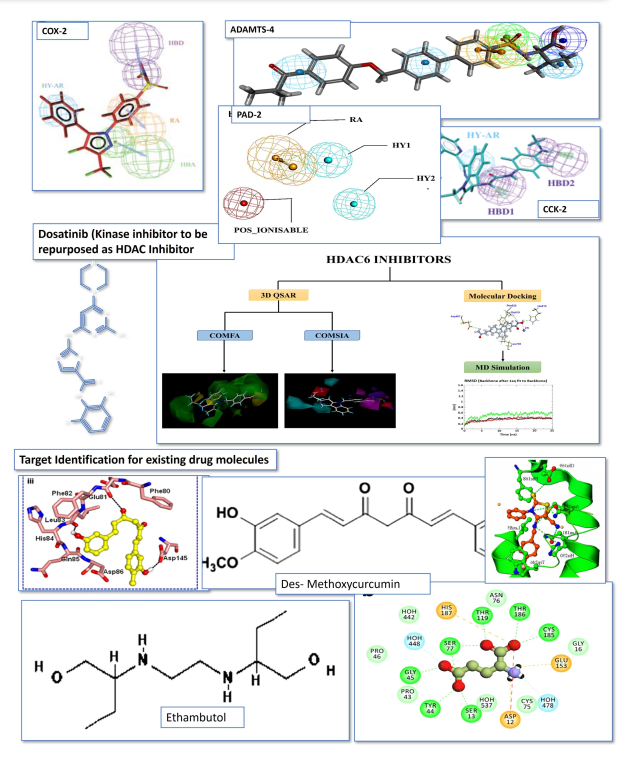
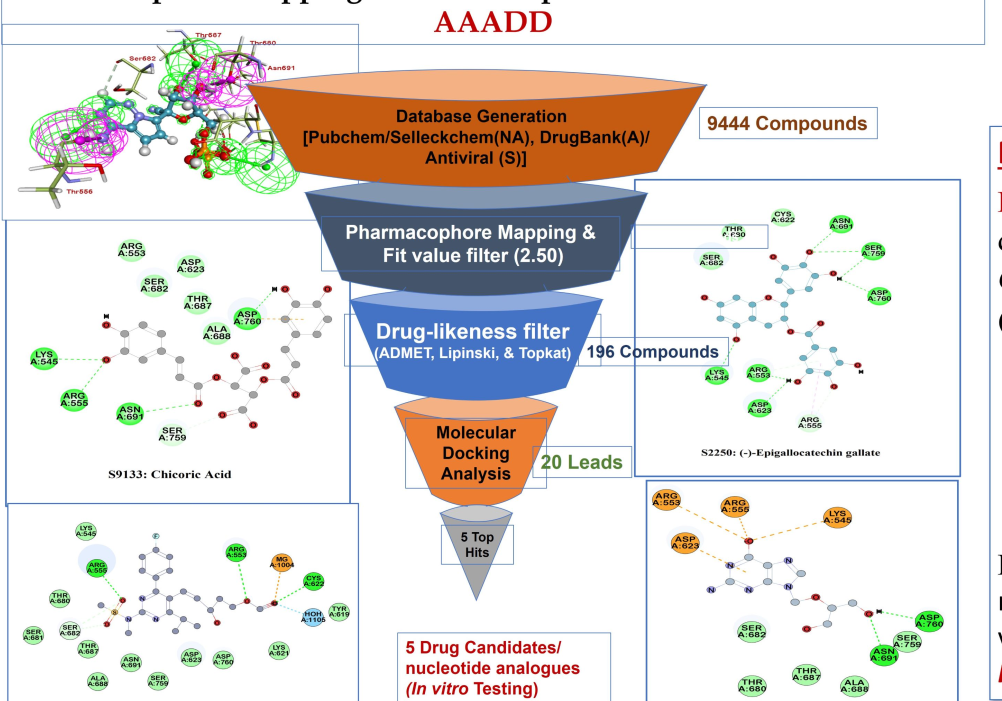
The Bioinformatics Center at ACBR, University of Delhi is dedicated to advancing tools, including computational ones, for the discovery of innovative drugs. The primary focus is on addressing the growing threat of infections and developing target-based cancer therapies. The center is actively engaged in research on identifying small molecules against various disease targets. The application of the PBVS approach has led to the discovery of potential HDAC6- specific inhibitors for cancer treatment and repurposed drugs for combatting SARS-CoV-2. This groundbreaking study significantly reduces both the time and financial resources required for discovering new molecules by leveraging computational approaches. Through in- silico methods, LY-2874455 and Pitavastatin have been identified as potential HDAC6-specific inhibitors, with their bioactivity against this new target validated through in-vitro evaluation in cell line-based models. Additionally, Chicoric acid, Ganciclovir, Lopinavir, Rosuvastatin, and Epigallate have been recognized as potential repurposed drugs acting as RdRp inhibitors for the treatment of SARS-CoV-2, with Chicoric acid and Epigallate demonstrating notable antiviral activity.
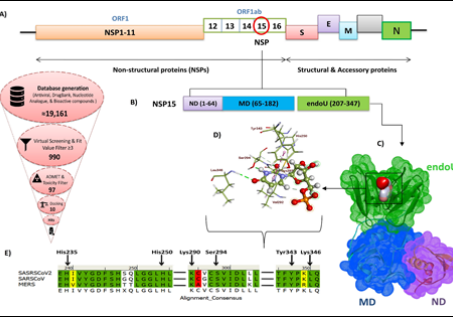
Moreover, the identification of Brivudine (S5009) as a lead for the Nsp15 of SARS-CoV-2 showcases the center's commitment to repurposing existing drugs for new applications. For phase I, five compounds (DB11781, S1639, S7259, S0391, and S1230) against the PanK target of MTb have been selected and will undergo in-vitro evaluation. The center has already published findings on some of the lead compounds discovered.
The center is also actively engaged in training of Ph.D., MSc, and other candidates annually. Equipped with a powerful computational facility, including the Param Shavak (HPC system), eight Xeon-based workstations, and specialized drug discovery software, the center is in the process of developing a drug discovery web server, ACBR_Ankalan. This server will predict the bioactivities of new molecules for anticancer activities and maintain databases of molecules with predicted activities. Leveraging molecular mechanics-based drug design along with modern ML and AI methods, the center addresses challenges in drug discovery and related biomedical sciences.
The ACBR center's broad research scope spans diseases caused by viruses, bacteria, and cancer. Their studies yield promising small molecules for potential drug development in these areas. By utilizing computational approaches, the center efficiently narrows down the pool of candidates, significantly reducing costs associated with drug discovery. In their commitment to combating diseases without cures, the center expedites the availability of new drugs to the public.
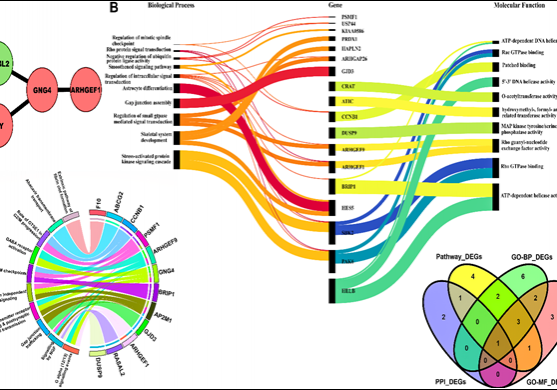
Furthermore, the ACBR center collaborates with IIIT-Delhi, INMAS, the Computer Science department, and other Delhi University colleges to integrate ML-AI into drug design and development. The center employs a genome-based systems biology approach and data mining to understand disease mechanisms and facilitate drug discovery. The ongoing collaborative project focuses on identifying lead compounds against cancer targets through an integrated approach involving machine learning and 3D QSAR/pharmacophore-based virtual screening. The results will be catalogued in databases like ACBR_AANKLAN. The systems biology approach, utilizing genome mining, aims to discover new targets for lifestyle/complex diseases, targeting epigenetic regulators, proteins, RNA molecules, and combating antimicrobial resistance.


