IISER, Mohali
Empowering Genomic and Proteomic Research: The Innovative Approach of the IISER Mohali
The Bioinformatics Center at IISER Mohali is dedicated to analyzing large-scale proteome and genomics data, with a focus on creating tools for enhancing bioinformatics education and facilitating research on large-scale genomic and proteome analyses. The advancement in high-throughput methodologies has resulted in vast genomic, epigenomic, and proteomic datasets, offering significant potential for systematic analysis to gain insights into biological processes and decipher molecular mechanisms underlying protein functions.
The center has specific objectives centered around harnessing extensive large-scale datasets. These objectives include creating the CBIOEduNet (Computational Biology Education network) suite, designed to provide comprehensive education and training programs. Additionally, the center aims to develop online teaching modules for students, post-doctoral fellows, and faculties. Priorities also include establishing the ExonVardb, a database dedicated to eukaryotic species' exons, alongside databases like Pepjunction, focusing on exon-exon junction peptides. The center further aims to create a database linking isoform heterogeneity with interaction partners and tissue specificity to deepen the understanding of biological dynamics. Additionally, the center intends to delve into high-throughput protein structure modeling, focusing on domain motions and their biological functions influenced by alternative splicing. It also aims to establish a robust server for processed datasets and in-house developed computer programs. Beyond these, the center plans to engage in research collaborations, offer associated services, and organize workshops and conferences to foster knowledge exchange and collaboration within the scientific community.


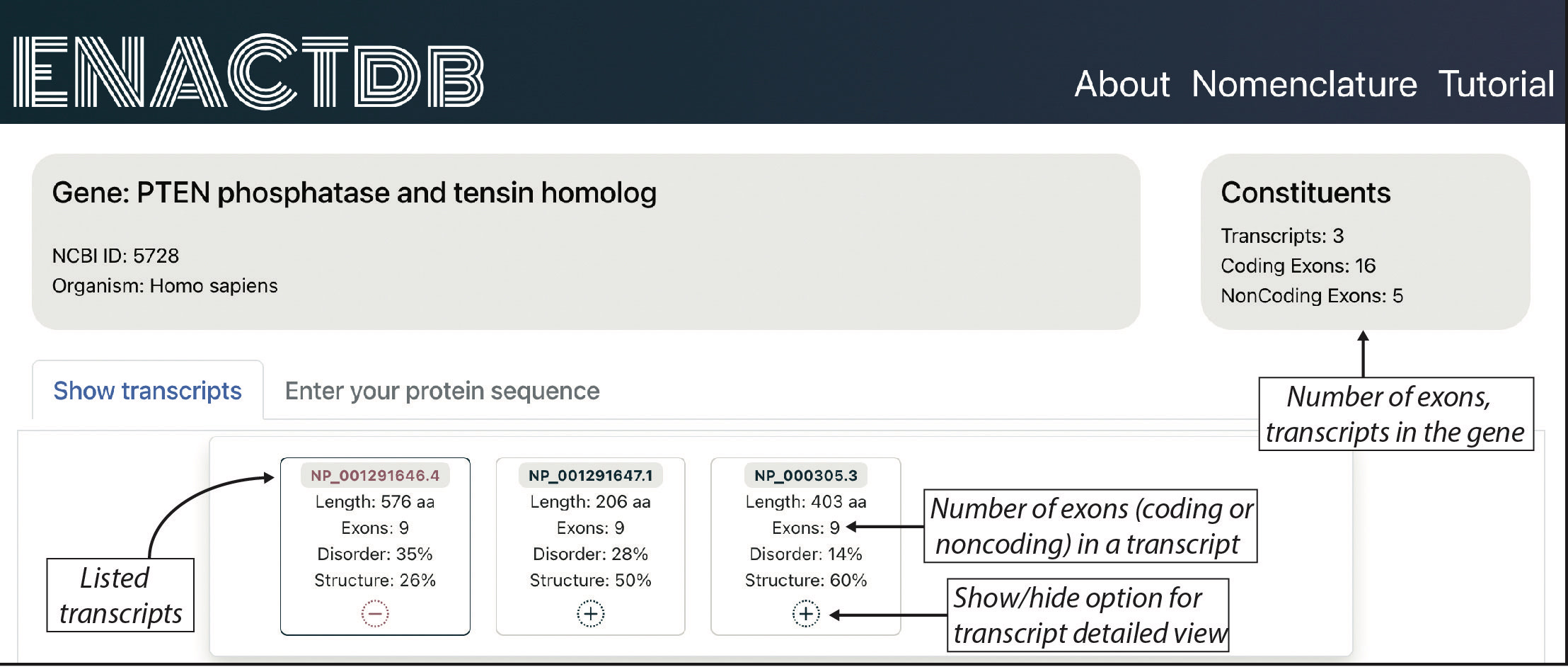
The center's achievements include the development of ENACTdb, a database compiling annotated exons from representative genomes to study the effect of alternative splicing on isoforms. The center has successfully organized a workshop for university lecturers and doctoral students, enhancing their proficiency in utilizing R, a crucial software in Bioinformatics analyses. Furthermore, the center has developed a unique nomenclature (ENACT) to annotate exons for analyzing isoform variations in alternative splicing. The annotated exons for five representative genomes have been documented in the ENACTdb database. Leveraging these annotated exons, the center pioneered a method using peptide junctions as signatures to identify protein isoform expression. Highlighting their expertise, the center adeptly handles high-throughput genomics data, performs diverse analyses, and conducts protein structural analyses to investigate its function.
One of the center's NNP partners is IHBT, Palampur, which has substantial expertise in developing RNAseq analysis tools to detect gene expression. Another NNP partner is the University of Hyderabad with expertise in gene regulation during cellular differentiation.
