NORTH-EASTERN HILL UNIVERSITY (NEHU), SHILLONG
Biodiversity Informatics: Unlocking the Genomic Secrets of Northeast India through Bioinformatics efforts at NEHU, Shillong
The Bioinformatics Center at NEHU, Shillong is devoted to Biodiversity Informatics, with a strong emphasis on critical research domains, including molecular characterization and genome diversity within orchid and carnivorous plant species. The center specializes in studying helminth parasites of medical and veterinary importance, particularly focusing on food-borne trematodiases. Its research further extends to exploring microbial diversity within mining areas and the sacred groves of Meghalaya.
North-east India holds exceptional biodiversity and serves as a pivotal gateway for India's rich flora and fauna, hosting a multitude of rare species. The spectrum includes traditional herbal drugs known for their pharmacognosy properties in treating severe illnesses to emerging infectious trematodiases, requiring immediate surveillance and control measures.


To accommodate the diverse spectrum of rare species and effectively address these challenges, the center has outlined specific objectives. The first objective focuses on enhancing E-infrastructure within Biodiversity Informatics, establishing important linkages and collaborations nationally and internationally to bolster biodiversity research activities. This center serves as a nodal center for the North-Eastern Bioinformatics Network (NEBINet). The second objective revolves around capacity building and covers organizing symposia, workshops, and brainstorming sessions at both national and international levels to promote industry-academia partnerships and strengthen scientific networks within the overarching theme of "Biodiversity Informatics". The third objective strives at training and skill development and involves initiating Bioinformatics@School to integrate research projects for high school students of different ages. Concurrently, to implement a teacher training program in Bioinformatics in partnership with the DBT-BIF Centers. Additionally, extending support to the NEHU INCUBATION CENTRE by facilitating bio-innovations, empowering young entrepreneurs, and fostering startups from the northeastern region.
The center’s accomplishments represent important milestones in biodiversity research. Notably, the sequencing and public release of the complete genome and transcriptome of the largest human intestinal fluke for the first time. Similarly, sequencing the organelles and the entire Indian pitcher plant's genome for the first time.
The establishment of the web-based portal "NEIHPID" showcasing taxonomical and NGS data on parasitic helminths serves as a cornerstone resource benefiting the research community and society at large. The knowledge base played a crucial role in unraveling the origins of food-borne tropical parasitic diseases essential for devising effective interventions and control strategies, profoundly influencing human well-being and societal welfare. Recognizing the evolving nature of sequencing methodologies, the center actively supports researchers in adopting cutting-edge techniques like Oxford Nanopore MinION Sequencing and Illumina. This endeavor includes workshops, biodiversity initiatives, software development, and expert speaker engagements, emphasizing the importance of northeastern biodiversity.
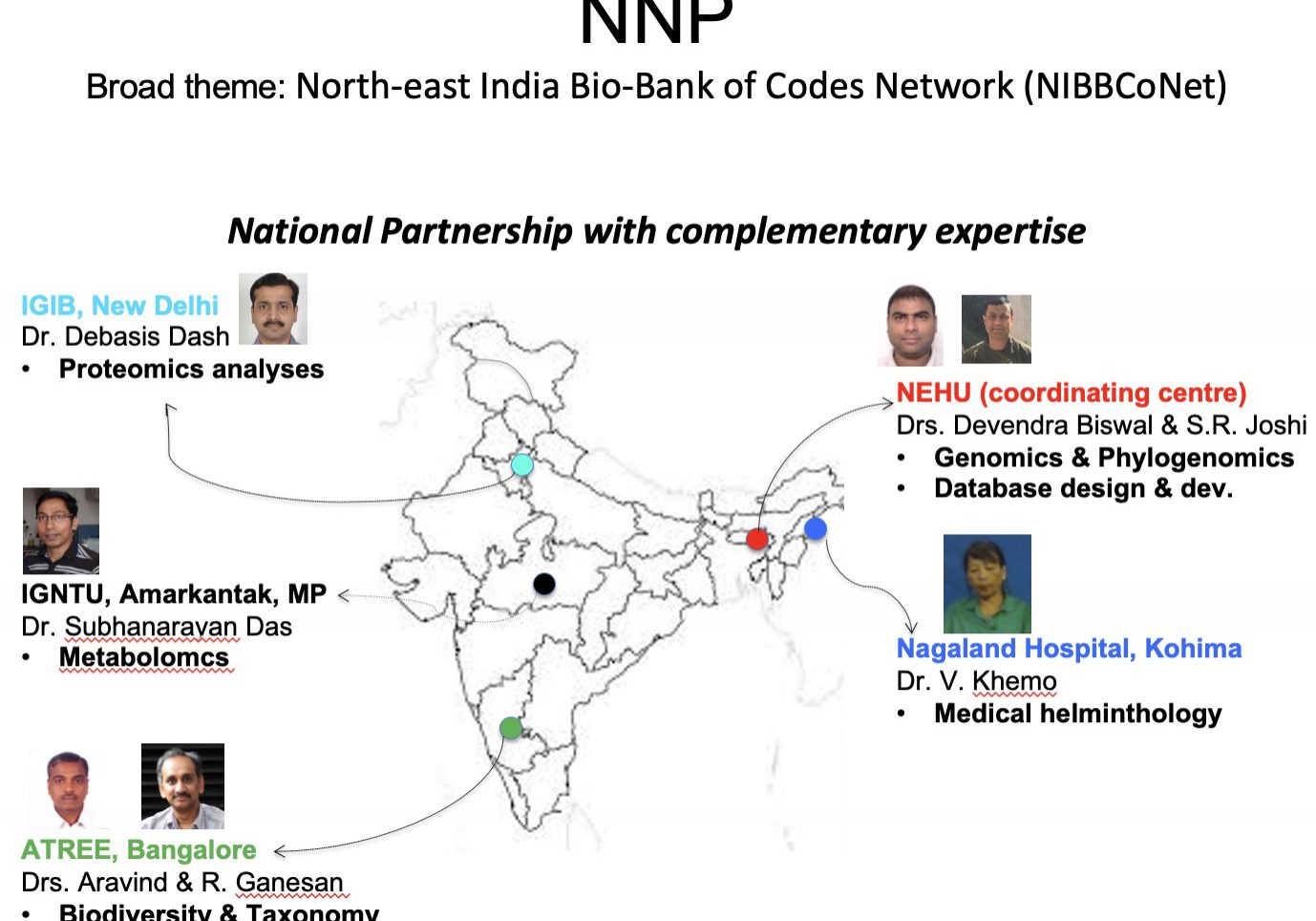
Under the NNP collaboration involving CSIR-IGIB, ATREE, IGNTU, the Healthcare Laboratory, and the Research Centre Naga Hospital Authority Kohima, BIC@NEHU is leading the generation of extensive and exceptional data in diverse realms. The focus is on Helminth parasite biodiversity, Orchid genomics, Microbial diversity sourced from mining sites, sacred grooves in the northeastern region, and carnivorous plants native to NER. The collaborative efforts will result in a wealth of high-quality and unique data, enriching the understanding of varied domains within biodiversity research.
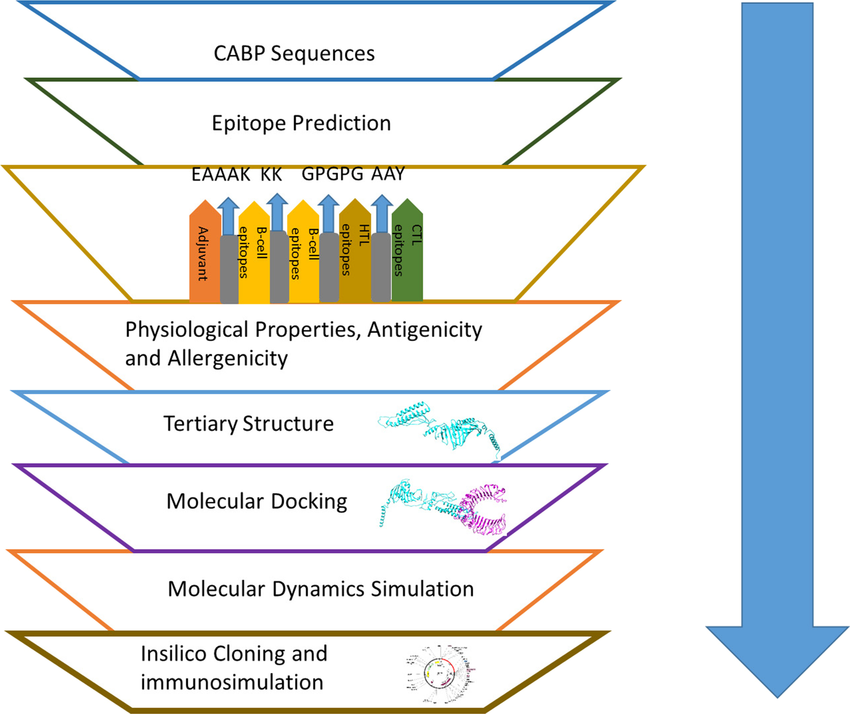
Schematic representation of the multi-epitope subunit vaccine candidate designing using B-cell, CTL and HTL epitopes followed by molecular docking, molecular dynamics simulation and in silico cloning.