Tamil Nadu Agricultural University (TNAU)
Bridging Agriculture and Bioinformatics: Innovations and Insights from TNAU Coimbatore
The Bioinformatics Center at TNAU Coimbatore is situated within the Department of Plant Molecular Biology and Bioinformatics at the Centre for Plant Molecular Biology and Biotechnology. The center's research primarily concentrates on millet crops, encompassing OMICS data analysis, allele mining for various agronomic traits, and the development of tools and databases. Its primary goal is to develop comprehensible computational approaches for analyzing plant life processes under diverse conditions to enhance sustainable agriculture.
The center has developed a crop genome database and FTP server, providing monthly updates for both newly sequenced and existing crop genomes. It endeavours to design and develop a Resource Description Framework (RDF) Search Engine, offering a unified user interface to interconnect diverse sources of crop genomic data. Additionally, it seeks to establish an automated workflow for key functions using next-generation sequencing data. The center aims to identify novel alleles associated with yield components, stress tolerance, grain quality, and nutrient use efficiency through genomics-assisted allele mining in crop plants, exemplified by rice and minor millets. Rice genotypes ASD16 and ADT43 with novel alleles affecting grain quality and starch content have been identified. An "A to G" polymorphism was found at the CDS position 220 of the An-1 gene, regulating awn formation and grain length. Non synonymous substitutions were also detected in starch biosynthesis genes. The center has developed a Next Generation Sequencing (NGS) data analysis pipeline prototype and a millet genomics database. A software package named NexaHub was created for NGS data analysis, covering quality controls through annotations, useful for researchers from diverse fields including molecular biologists and those less familiar with programming. They also developed a Comprehensive Millet Omics Resource named MIMI DB.
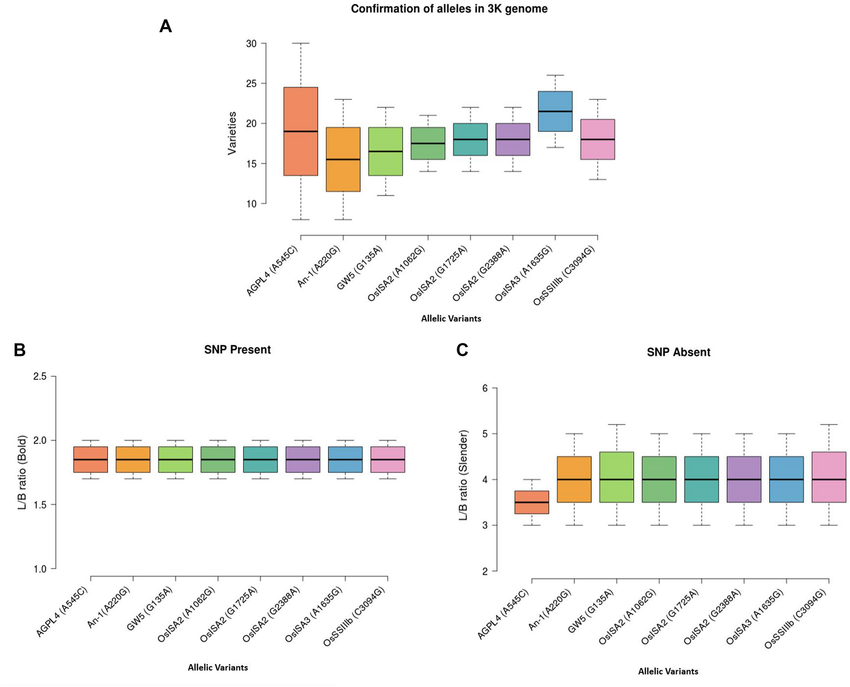
Validation of allelic variants of grain size and starch biosynthesis-related genes from ASD16 and ADT43 and their level of confirmation in 3K genome data.
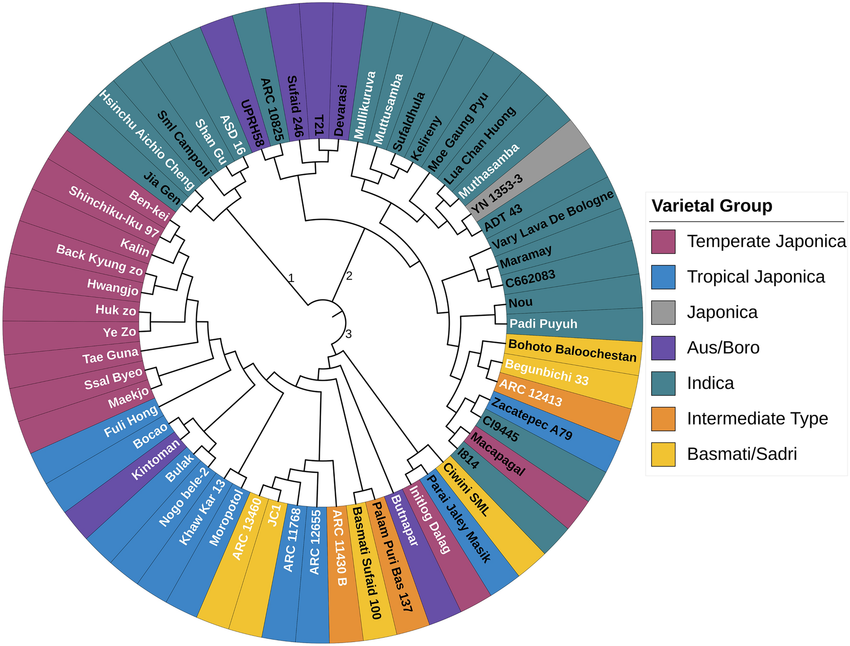
Dendrogram showing the evolutionary relationship between the rice genotypes. The ADT43 is closely related to YN 1353-3 and ASD16 is closely related to Shan Gu. The white colour font labels indicate the bold grain type and the black colour font labels indicate the slender grain type. The different background colours of the labels indicate varietal groups.
Furthermore, the center has filed copyrights for various in-house developed software and databases, such as MaizeSSRdb, VignaGuard, MIMI DB – An integrated multi-omics database on Minor Millets, and NexaHub: NGS Data analysis pipeline. The center actively engaged in providing training, short courses, workshops, and symposiums in bioinformatics, along with launching a new master program (Figure 5). A virtual workshop on computational genomics was also organized, attracting 50 participants.
As part of the National Network Project, the center with its collaborating parters IIT-Madras, The Central University of Karnataka, and ICAR-NBAIR, Bengaluru are dedicated to managing Papaya Ring Spot Virus through molecular approaches, deciphering genes and bioactive molecules from Solanaceae species, and understanding the microbiome of termitarium and termite gut. The project aims to target complex protein-protein interactions to impede pathogen transmission and develop safe phyto-molecule inhibitor formulations. Using interdisciplinary approaches, including nanotechnology, discovery of novel biomolecules, and metagenomic deciphering of termite functional genes, the project aims to analyse therapeutic, insecticidal, and antimicrobial properties in Solanaceae species, along with investigating cultivable microbial diversity for potential biotechnological applications.

The center has organized various hands on training, short courses, workshops, and symposiums in NGS data analysis, Molecular modeling, docking and simulation and RNA-seq assembly and annotation.

